An introduction to Scikit-Learn | Get Instant Help In Machine Learning
- realcode4you
- Jun 4, 2021
- 5 min read
Examples require a Python distribution with scientific packages:
Jump to https://www.continuum.io/downloads and download the installer.
bash Anaconda2-4.2.0-Linux-x86_64.sh (or whatever installer you picked)
conda install scikit-learn numpy scipy matplotlib jupyter pandas
You are ready to go!
# Global imports and settings
# Matplotlib
%matplotlib inline
from matplotlib import pyplot as plt
plt.rcParams["figure.max_open_warning"] = -1
# Print options
import numpy as np
np.set_printoptions(precision=3)
# Slideshow
from notebook.services.config import ConfigManager
cm = ConfigManager()
cm.update('livereveal', {'width': 1440, 'height': 768, 'scroll': True, 'theme': 'simple'})
# Silence warnings
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=UserWarning)
warnings.simplefilter(action="ignore", category=RuntimeWarning)Outline
Scikit-Learn and the scientific ecosystem in Python
Supervised learning
Transformers, pipelines and feature unions
Beyond building classifiers
Summary
Scikit-Learn
Machine learning library written in Python
Simple and efficient, for both experts and non-experts
Classical, well-established machine learning algorithms
Shipped with documentation and examples
BSD 3 license
Python stack for data analysis
The open source Python ecosystem provides a standalone, versatile and powerful scientific working environment, including: NumPy, SciPy, Jupyter Matplotlib, Pandas, and many others...

Scikit-Learn builds upon NumPy and SciPy and complements this scientific environment with machine learning algorithms;
By design, Scikit-Learn is non-intrusive, easy to use and easy to combine with other libraries;
Core algorithms are implemented in low-level languages.
Algorithms
Supervised learning:
Linear models (Ridge, Lasso, Elastic Net, ...)
Support Vector Machines
Tree-based methods (Random Forests, Bagging, GBRT, ...)
Nearest neighbors
Neural networks
Gaussian Processes
Feature selection
Unsupervised learning:
Clustering (KMeans, Ward, ...)
Matrix decomposition (PCA, ICA, ...)
Density estimation
Outlier detection
Model selection and evaluation:
Cross-validation
Grid-search
Lots of metrics
Supervised learning



Applications
Classifying signal from background events;
Diagnosing disease from symptoms;
Recognising cats in pictures;
Identifying body parts with cameras;
Predicting temperature for the next days
Data
Input data = Numpy arrays or Scipy sparse matrices ;
Algorithms are expressed using high-level operations defined on matrices or vectors (similar to MATLAB) ;
Leverage efficient low-leverage implementations ;
Keep code short and readable.
# Generate data
from sklearn.datasets import make_blobs
X, y = make_blobs(n_samples=300, centers=20, random_state=123)
labels = ["b", "r"]
y = np.take(labels, (y < 10))
print(X[:5])
print(y[:5])
Output:
[[-2.956 -3.749]
[-7.586 2.066]
[ 0.457 8.059]
[-5.996 2.021]
[-0.979 -9.781]]
['b' 'r' 'b' 'b' 'r']
#Print the shape
# X is a 2 dimensional array, with 300 rows and 2 columns
print(X.shape)
# y is a vector of 300 elements
print(y.shape)
Result:
(300, 2) (300,)
Accessing row and column
# Rows and columns can be accessed with lists, slices or masks
print(X[[1, 2, 3]]) # rows 1, 2 and 3
print(X[:5]) # 5 first rows
print(X[200:210, 0]) # values from row 200 to row 210 at column 0
print(X[y == "b"][:5]) # 5 first rows for which y is "b"Output:
[[-7.586 2.066]
[ 0.457 8.059]
[-5.996 2.021]]
[[-2.956 -3.749]
[-7.586 2.066]
[ 0.457 8.059]
[-5.996 2.021]
[-0.979 -9.781]]
[ -1.448 -6.3 -6.195 -1.99 -3.411 -7.009 5.402 -4.995 10.883
-6.661]
[[-2.956 -3.749]
[ 0.457 8.059]
[-5.996 2.021]
[-4.021 -5.173]
[ 4.01 2.581]]# Plot
for label in labels:
mask = (y == label)
plt.scatter(X[mask, 0], X[mask, 1], c=label, linewidths=0)
plt.xlim(-10, 10)
plt.ylim(-10, 10)
plt.show()Output

A simple and unified API
All learning algorithms in scikit-learn share a uniform and limited API consisting of complementary interfaces:
an estimator interface for building and fitting models;
a predictor interface for making predictions;
a transformer interface for converting data.
Goal: enforce a simple and consistent API to make it trivial to swap or plug algorithms.

Estimators
class Estimator(object):
def fit(self, X, y=None):
"""Fits estimator to data."""
# set state of ``self``
# ...
return self# Import the nearest neighbor class
from sklearn.neighbors import KNeighborsClassifier # Change this to try
# something else
# Set hyper-parameters, for controlling algorithm
clf = KNeighborsClassifier(n_neighbors=5)
# Learn a model from training data
clf.fit(X, y)Result
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform')
# Estimator state is stored in instance attributes
clf._treeOutput
<sklearn.neighbors.kd_tree.KDTree at 0x33ecde8>
Predictors
# Make predictions
print(clf.predict(X[:5])) result:
['b' 'r' 'b' 'b' 'r']
# Compute (approximate) class probabilities
print(clf.predict_proba(X[:5]))result:
[[ 1. 0. ] [ 0.4 0.6] [ 1. 0. ] [ 0.6 0.4] [ 0. 1. ]]
from tutorial import plot_surface
plot_surface(clf, X, y)result:

from tutorial import plot_histogram
plot_histogram(clf, X, y)
Classifier zoo
Decision trees
Idea: greedily build a partition of the input space using cuts orthogonal to feature axes.
from tutorial import plot_clf
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier()
clf.fit(X, y)
plot_clf(clf, X, y)result:

Random Forests
Idea: Build several decision trees with controlled randomness and average their decisions.
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=500)
# from sklearn.ensemble import ExtraTreesClassifier
# clf = ExtraTreesClassifier(n_estimators=500)
clf.fit(X, y)
plot_clf(clf, X, y)result:

Support vector machines
Idea: Find the hyperplane which has the largest distance to the nearest training points of any class.
from sklearn.svm import SVC
clf = SVC(kernel="linear") # try kernel="rbf" instead
clf.fit(X, y)
plot_clf(clf, X, y)result

Multi-layer perceptron
Idea: a multi-layer perceptron is a circuit of non-linear combinations of the data.
from sklearn.neural_network import MLPClassifier
clf = MLPClassifier(hidden_layer_sizes=(100, 100, 100),
activation="tanh",
learning_rate="invscaling")
clf.fit(X, y)
plot_clf(clf, X, y)result:

Gaussian Processes
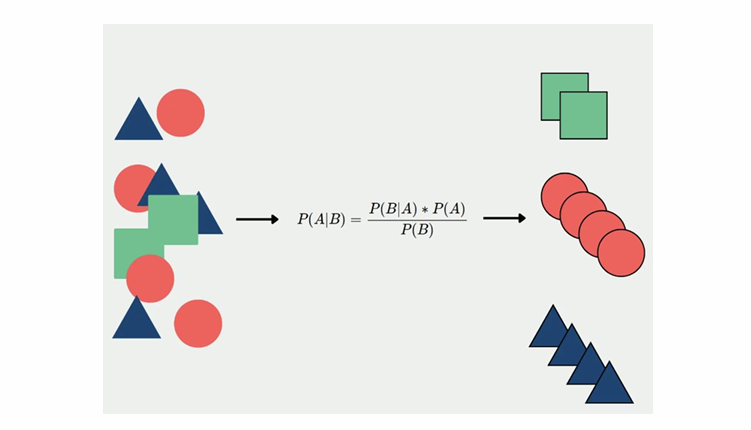
Idea: a gaussian process is a distribution over functions 𝑓f, such that 𝑓(𝐱)f(x), for any set 𝐱x of points, is gaussian distributed.
from sklearn.gaussian_process import GaussianProcessClassifier
clf = GaussianProcessClassifier()
clf.fit(X, y)
plot_clf(clf, X, y)
result:

Transformers, pipelines and feature unions
Transformers
Classification (or regression) is often only one or the last step of a long and complicated process;
In most cases, input data needs to be cleaned, massaged or extended before being fed to a learning algorithm;
For this purpose, Scikit-Learn provides the transformer API.
class Transformer(object):
def fit(self, X, y=None):
"""Fits estimator to data."""
# set state of ``self``
return self
def transform(self, X):
"""Transform X into Xt."""
# transform X in some way to produce Xt
return Xt
# Shortcut
def fit_transform(self, X, y=None):
self.fit(X, y)
Xt = self.transform(X)
return Xt
Transformer zoo
# Load digits data
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
digits = load_digits()
X, y = digits.data, digits.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Plot
sample_id = 42
plt.imshow(X[sample_id].reshape((8, 8)), interpolation="nearest", cmap=plt.cm.Blues)
plt.title("y = %d" % y[sample_id])
plt.show()
result:

Scalers and other normalizers
from sklearn.preprocessing import StandardScaler
tf = StandardScaler()
tf.fit(X_train, y_train)
Xt_train = tf.transform(X_train)
print("Mean (before scaling) =", np.mean(X_train))
print("Mean (after scaling) =", np.mean(Xt_train))
# Shortcut: Xt = tf.fit_transform(X)
# See also Binarizer, MinMaxScaler, Normalizer, ...result:
Mean (before scaling) = 4.89212138085
Mean (after scaling) = -2.30781326574e-18# Scaling is critical for some algorithms
from sklearn.svm import SVC
clf = SVC()
print("Without scaling =", clf.fit(X_train, y_train).score(X_test, y_test))
print("With scaling =", clf.fit(tf.transform(X_train), y_train).score(tf.transform(X_test), y_test))result:
Without scaling = 0.486666666667
With scaling = 0.984444444444Feature selection
# Select the 10 top features, as ranked using ANOVA F-score
from sklearn.feature_selection import SelectKBest, f_classif
tf = SelectKBest(score_func=f_classif, k=10)
Xt = tf.fit_transform(X_train, y_train)
print("Shape =", Xt.shape)
# Plot support
plt.imshow(tf.get_support().reshape((8, 8)), interpolation="nearest", cmap=plt.cm.Blues)
plt.show()
Shape = (1347, 10)
result:

Feature selection (cont.)
# Feature selection using backward elimination
from sklearn.feature_selection import RFE
from sklearn.ensemble import RandomForestClassifier
tf = RFE(RandomForestClassifier(), n_features_to_select=10, verbose=1)
Xt = tf.fit_transform(X_train, y_train)
print("Shape =", Xt.shape)
# Plot support
plt.imshow(tf.get_support().reshape((8, 8)), interpolation="nearest", cmap=plt.cm.Blues)
plt.show()
result:
Fitting estimator with 64 features.
Fitting estimator with 63 features.
Fitting estimator with 62 features.
Fitting estimator with 61 features.
Fitting estimator with 60 features.
Fitting estimator with 59 features.
Fitting estimator with 58 features.
Fitting estimator with 57 features.
Fitting estimator with 56 features.
Fitting estimator with 55 features.
Fitting estimator with 54 features.
Fitting estimator with 53 features.
Fitting estimator with 52 features.
Fitting estimator with 51 features.
Fitting estimator with 50 features.
Fitting estimator with 49 features.
------
Decomposition, factorization or embeddings
# Compute decomposition
# from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
tf = TSNE(n_components=2)
Xt_train = tf.fit_transform(X_train)
# Plot
plt.scatter(Xt_train[:, 0], Xt_train[:, 1], c=y_train, linewidths=0)
plt.show()
# See also: KernelPCA, NMF, FastICA, Kernel approximations,
# manifold learning, etc
Function transformer
from sklearn.preprocessing import FunctionTransformer
def increment(X):
return X + 1
tf = FunctionTransformer(func=increment)
Xt = tf.fit_transform(X)
print(X[0])
print(Xt[0])Output:
[ 0. 0. 5. 13. 9. 1. 0. 0. 0. 0. 13. 15. 10. 15. 5.
0. 0. 3. 15. 2. 0. 11. 8. 0. 0. 4. 12. 0. 0. 8.
8. 0. 0. 5. 8. 0. 0. 9. 8. 0. 0. 4. 11. 0. 1.
12. 7. 0. 0. 2. 14. 5. 10. 12. 0. 0. 0. 0. 6. 13.
10. 0. 0. 0.]
[ 1. 1. 6. 14. 10. 2. 1. 1. 1. 1. 14. 16. 11. 16. 6.
1. 1. 4. 16. 3. 1. 12. 9. 1. 1. 5. 13. 1. 1. 9.
9. 1. 1. 6. 9. 1. 1. 10. 9. 1. 1. 5. 12. 1. 2.
13. 8. 1. 1. 3. 15. 6. 11. 13. 1. 1. 1. 1. 7. 14.
11. 1. 1. 1.]Send your request to get any other machine learning assignment or project help using sklearn or from sketch at realcode4you@gmail.com


Comments