Pytorch Assignment Help, Project Help and Homework Help | Introduction to the Pytorch
- realcode4you
- Feb 26, 2023
- 11 min read
Introduction
PyTorch is a machine learning framework that is used in both academia and industry for various applications. PyTorch started of as a more flexible alternative to TensorFlow, which is another popular machine learning framework. At the time of its release, PyTorch appealed to the users due to its user friendly nature: as opposed to defining static graphs before performing an operation as in TensorFlow, PyTorch allowed users to define their operations as they go, which is also the approached integrated by TensorFlow in its following releases. Although TensorFlow is more widely preferred in the industry, PyTorch is often times the preferred machine learning framework for researchers. If you would like to learn more about the differences between the two, you can check out this blog post.
Now that we have learned enough about the background of PyTorch, let's start by importing it into our notebook. To install PyTorch, you can follow the instructions here. Alternatively, you can open this notebook using Google Colab, which already has PyTorch installed in its base kernel. Once you are done with the installation process, run the following cell:
import torch
import torch.nn as nn
# Import pprint, module we use for making our print statements prettier
import pprint
pp = pprint.PrettyPrinter()We are all set to start our tutorial. Let's dive in!
Tensors Tensors are the most basic building blocks in PyTorch. Tensors are similar to matrices, but the have extra properties and they can represent higher dimensions. For example, an square image with 256 pixels in both sides can be represented by a 3x256x256 tensor, where the first 3 dimensions represent the color channels, red, green and blue.
Tensor Initialization There are several ways to instantiate tensors in PyTorch, which we will go through next.
From a Python List We can initalize a tensor from a Python list, which could include sublists. The dimensions and the data types will be automatically inferred by PyTorch when we use torch.tensor()
# Initialize a tensor from a Python List
data = [
[0, 1],
[2, 3],
[4, 5]
]
x_python = torch.tensor(data)
# Print the tensor
x_pythonOutput:
tensor([[0, 1], [2, 3], [4, 5]])
We can also call torch.tensor() with the optional dtype parameter, which will set the data type. Some useful datatypes to be familiar with are: torch.bool, torch.float, and torch.long.
# We are using the dtype to create a tensor of particular type
x_float = torch.tensor(data, dtype=torch.float)
x_floatOutput:
tensor([[0., 1.], [2., 3.], [4., 5.]])
# We are using the dtype to create a tensor of particular type
x_bool = torch.tensor(data, dtype=torch.bool)
x_boolOutput:
tensor([[False, True], [ True, True], [ True, True]])
We can also get the same tensor in our specified data type using methods such as float(), long() etc.
x_python.float()output:
tensor([[0., 1.], [2., 3.], [4., 5.]])
We can also use tensor.FloatTensor, tensor.LongTensor, tensor.Tensor classes to instantiate a tensor of particular type. LongTensors are particularly important in NLP as many methods that deal with indices require the indices to be passed as a LongTensor, which is a 64 bit integer.
# `torch.Tensor` defaults to float
# Same as torch.FloatTensor(data)
x = torch.Tensor(data)
xoutput:
tensor([[0., 1.], [2., 3.], [4., 5.]])
From a NumPy Array We can also initialize a tensor from a NumPy array.
import numpy as np
# Initialize a tensor from a NumPy array
ndarray = np.array(data)
x_numpy = torch.from_numpy(ndarray)
# Print the tensor
x_numpyoutput:
tensor([[0, 1], [2, 3], [4, 5]])
From a Tensor We can also initialize a tensor from another tensor, using the following methods:
torch.ones_like(old_tensor): Initializes a tensor of 1s.
torch.zeros_like(old_tensor): Initializes a tensor of 0s.
torch.rand_like(old_tensor): Initializes a tensor where all the elements are sampled from a uniform distribution between 0 and 1.
torch.randn_like(old_tensor): Initializes a tensor where all the elements are sampled from a normal distribution.
All of these methods preserve the tensor properties of the original tensor passed in, such as the shape and device, which we will cover in a bit.
# Initialize a base tensor
x = torch.tensor([[1., 2], [3, 4]])
xoutput:
tensor([[1., 2.], [3., 4.]])
# Initialize a tensor of 0s
x_zeros = torch.zeros_like(x)
x_zerosoutput:
tensor([[0., 0.], [0., 0.]])
# Initialize a tensor of 1s
x_ones = torch.ones_like(x)
x_onesoutput:
tensor([[1., 1.], [1., 1.]])
# Initialize a tensor where each element is sampled from a uniform distribution
# between 0 and 1
x_rand = torch.rand_like(x)
x_randoutput:
tensor([[0.8979, 0.7173], [0.3067, 0.1246]])
# Initialize a tensor where each element is sampled from a normal distribution
x_randn = torch.randn_like(x)
x_randnoutput:
tensor([[-0.6749, -0.8590], [ 0.6666, 1.1185]])
By Specifying a Shape We can also instantiate tensors by specifying their shapes (which we will cover in more detail in a bit). The methods we could use follow the ones in the previous section:
torch.zeros()
torch.ones()
torch.rand()
torch.randn()
# Initialize a 2x3x2 tensor of 0s
shape = (4, 2, 2)
x_zeros = torch.zeros(shape) # x_zeros = torch.zeros(4, 3, 2) is an alternative
x_zerosoutput:
tensor([[[0., 0.], [0., 0.]], [[0., 0.], [0., 0.]], [[0., 0.], [0., 0.]], [[0., 0.], [0., 0.]]])
With torch.arange() We can also create a tensor with torch.arange(end), which returns a 1-D tensor with elements ranging from 0 to end-1. We can use the optional start and step parameters to create tensors with different ranges.
# Create a tensor with values 0-9
x = torch.arange(10)
xOutput:
tensor([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
Tensor Properties Tensors have a few properties that are important for us to cover. These are namely shape, and the device properties.
Data Type The dtype property lets us see the data type of a tensor.
# Initialize a 3x2 tensor, with 3 rows and 2 columns
x = torch.ones(3, 2)
x.dtypeOutput:
torch.float32
Shape The shape property tells us the shape of our tensor. This can help us identify how many dimensional our tensor is as well as how many elements exist in each dimension.
# Initialize a 3x2 tensor, with 3 rows and 2 columns
x = torch.Tensor([[1, 2], [3, 4], [5, 6]])
xOutput:
tensor([[1., 2.], [3., 4.], [5., 6.]])
# Print out its shape
# Same as x.size()
x.shapeOutput:
torch.Size([3, 2])
# Print out the number of elements in a particular dimension
# 0th dimension corresponds to the rows
x.shape[0]Output:
3
We can also get the size of a particular dimension with the size() method.
# Get the size of the 0th dimension
x.size(0)output:
3
We can change the shape of a tensor with the view() method.
# Example use of view()
# x_view shares the same memory as x, so changing one changes the other
x_view = x.view(2, 3)
x_viewoutput:
tensor([[1., 2., 3.], [4., 5., 6.]])
# We can ask PyTorch to infer the size of a dimension with -1
x_view = x.view(3, -1)
x_viewOutput:
tensor([[1., 2.], [3., 4.], [5., 6.]])
We can also use torch.reshape() method for a similar purpose. There is a subtle difference between reshape() and view(): view() requires the data to be stored contiguously in the memory. You can refer to this StackOverflow answer for more information. In simple terms, contiguous means that the way our data is laid out in the memory is the same as the way we would read elements from it. This happens because some methods, such as transpose() and view(), do not actually change how our data is stored in the memory. They just change the meta information about out tensor, so that when we use it we will see the elements in the order we expect.
reshape() calls view() internally if the data is stored contiguously, if not, it returns a copy. The difference here isn't too important for basic tensors, but if you perform operations that make the underlying storage of the data non-contiguous (such as taking a transpose), you will have issues using view(). If you would like to match the way your tensor is stored in the memory to how it is used, you can use the contiguous() method.
# Change the shape of x to be 3x2
# x_reshaped could be a reference to or copy of x
x_reshaped = torch.reshape(x, (2, 3))
x_reshapedOutput:
tensor([[1., 2., 3.], [4., 5., 6.]])
We can use torch.unsqueeze(x, dim) function to add a dimension of size 1 to the provided dim, where x is the tensor. We can also use the corresponding use torch.squeeze(x), which removes the dimensions of size 1.
# Initialize a 5x2 tensor, with 5 rows and 2 columns
x = torch.arange(10).reshape(5, 2)
xOutput:
tensor([[0, 1], [2, 3], [4, 5], [6, 7], [8, 9]])
# Add a new dimension of size 1 at the 1st dimension
x = x.unsqueeze(1)
x.shapeoutput:
torch.Size([5, 1, 2])
# Squeeze the dimensions of x by getting rid of all the dimensions with 1 element
x = x.squeeze()
x.shapeOutput:
torch.Size([5, 2])
If we want to get the total number of elements in a tensor, we can use the numel() method.
xOutput:
tensor([[0, 1], [2, 3], [4, 5], [6, 7], [8, 9]])
# Get the number of elements in tensor.
x.numel()output:
10Device Device property tells PyTorch where to store our tensor. Where a tensor is stored determines which device, GPU or CPU, would be handling the computations involving it. We can find the device of a tensor with the device property.
# Initialize an example tensor
x = torch.Tensor([[1, 2], [3, 4]])
xOutput:
tensor([[1., 2.], [3., 4.]])
# Get the device of the tensor
x.deviceoutput:
device(type='cpu')
We can move a tensor from one device to another with the method to(device).
# Check if a GPU is available, if so, move the tensor to the GPU
if torch.cuda.is_available():
x.to('cuda') Tensor Indexing In PyTorch we can index tensors, similar to NumPy.
# Initialize an example tensor
x = torch.Tensor([
[[1, 2], [3, 4]],
[[5, 6], [7, 8]],
[[9, 10], [11, 12]]
])
xoutput:
tensor([[[ 1., 2.],
[ 3., 4.]],
[[ 5., 6.],
[ 7., 8.]],
[[ 9., 10.],
[11., 12.]]])x.shapeoutput:
torch.Size([3, 2, 2])
# Access the 0th element, which is the first row
x[0] # Equivalent to x[0, :]output:
tensor([[1., 2.], [3., 4.]])
We can also index into multiple dimensions with :.
# Get the top left element of each element in our tensor
x[:, 0, 0]output:
tensor([1., 5., 9.])
Operations PyTorch operations are very similar to those of NumPy. We can work with both scalars and other tensors.
# Create an example tensor
x = torch.ones((3,2,2))
xoutput:
tensor([[[1., 1.], [1., 1.]], [[1., 1.], [1., 1.]], [[1., 1.], [1., 1.]]])
# Perform elementwise addition
# Use - for subtraction
x + 2output:
tensor([[[3., 3.], [3., 3.]], [[3., 3.], [3., 3.]], [[3., 3.], [3., 3.]]])
# Perform elementwise multiplication
# Use / for division
x * 2output:
tensor([[[2., 2.], [2., 2.]], [[2., 2.], [2., 2.]], [[2., 2.], [2., 2.]]])
We can apply the same operations between different tensors of compatible sizes.
# Create a 4x3 tensor of 6s
a = torch.ones((4,3)) * 6
aoutput:
tensor([[6., 6., 6.], [6., 6., 6.], [6., 6., 6.], [6., 6., 6.]])
# Create a 1D tensor of 2s
b = torch.ones(3) * 2
boutput:
tensor([2., 2., 2.])
# Divide a by b
a / boutput:
tensor([[3., 3., 3.], [3., 3., 3.], [3., 3., 3.], [3., 3., 3.]])
Autograd PyTorch and other machine learning libraries are known for their automatic differantiation feature. That is, given that we have defined the set of operations that need to be performed, the framework itself can figure out how to compute the gradients. We can call the backward() method to ask PyTorch to calculate the gradiends, which are then stored in the grad attribute.
# Create an example tensor
# requires_grad parameter tells PyTorch to store gradients
x = torch.tensor([2.], requires_grad=True)
# Print the gradient if it is calculated
# Currently None since x is a scalar
pp.pprint(x.grad)Output:
None
# Calculating the gradient of y with respect to x
y = x * x * 3 # 3x^2
y.backward()
pp.pprint(x.grad) # d(y)/d(x) = d(3x^2)/d(x) = 6x = 12output:
tensor([12.])
Let's run backprop from a different tensor again to see what happens.
z = x * x * 3 # 3x^2
z.backward()
pp.pprint(x.grad)output:
tensor([48.])
We can see that the x.grad is updated to be the sum of the gradients calculated so far. When we run backprop in a neural network, we sum up all the gradients for a particular neuron before making an update. This is exactly what is happening here! This is also the reason why we need to run zero_grad() in every training iteration (more on this later). Otherwise our gradients would keep building up from one training iteration to the other, which would cause our updates to be wrong.
Neural Network Module So far we have looked into the tensors, their properties and basic operations on tensors. These are especially useful to get familiar with if we are building the layers of our network from scratch. We will utilize these in Assignment 3, but moving forward, we will use predefined blocks in the torch.nn module of PyTorch. We will then put together these blocks to create complex networks. Let's start by importing this module with an alias so that we don't have to type torch every time we use it.
import torch.nn as nnLinear Layer We can use nn.Linear(H_in, H_out) to create a a linear layer. This will take a matrix of (N, *, H_in) dimensions and output a matrix of (N, *, H_out). The * denotes that there could be arbitrary number of dimensions in between. The linear layer performs the operation Ax+b, where A and b are initialized randomly. If we don't want the linear layer to learn the bias parameters, we can initialize our layer with bias=False.
# Create the inputs
input = torch.ones(2,3,4)
# N* H_in -> N*H_out
# Make a linear layers transforming N,*,H_in dimensinal inputs to N,*,H_out
# dimensional outputs
linear = nn.Linear(4, 2)
nn.Linear(2,1)
linear_output = linear(input)
linear_outputoutput:
tensor([[[-0.0935, 0.6382],
[-0.0935, 0.6382],
[-0.0935, 0.6382]],
[[-0.0935, 0.6382],
[-0.0935, 0.6382],
[-0.0935, 0.6382]]], grad_fn=<AddBackward0>)list(linear.parameters()) # Ax + boutput:
[Parameter containing: tensor([[-0.2491, 0.2283, 0.2765, -0.4489], [ 0.3642, 0.0685, -0.3154, 0.2699]], requires_grad=True), Parameter containing: tensor([0.0997, 0.2510], requires_grad=True)]
Other Module Layers There are several other preconfigured layers in the nn module. Some commonly used examples are nn.Conv2d, nn.ConvTranspose2d, nn.BatchNorm1d, nn.BatchNorm2d, nn.Upsample and nn.MaxPool2d among many others. We will learn more about these as we progress in the course. For now, the only important thing to remember is that we can treat each of these layers as plug and play components: we will be providing the required dimensions and PyTorch will take care of setting them up.
Activation Function Layer We can also use the nn module to apply activations functions to our tensors. Activation functions are used to add non-linearity to our network. Some examples of activations functions are nn.ReLU(), nn.Sigmoid() and nn.LeakyReLU(). Activation functions operate on each element seperately, so the shape of the tensors we get as an output are the same as the ones we pass in.
linear_outputoutput:
tensor([[[-0.0935, 0.6382],
[-0.0935, 0.6382],
[-0.0935, 0.6382]],
[[-0.0935, 0.6382],
[-0.0935, 0.6382],
[-0.0935, 0.6382]]], grad_fn=<AddBackward0>)sigmoid = nn.Sigmoid()
output = sigmoid(linear_output)
outputoutput:
tensor([[[0.4766, 0.6543], [0.4766, 0.6543], [0.4766, 0.6543]], [[0.4766, 0.6543], [0.4766, 0.6543], [0.4766, 0.6543]]], grad_fn=<SigmoidBackward>)
Putting the Layers Together So far we have seen that we can create layers and pass the output of one as the input of the next. Instead of creating intermediate tensors and passing them around, we can use nn.Sequentual, which does exactly that.
block = nn.Sequential(
nn.Linear(4, 2),
nn.Sigmoid()
)
input = torch.ones(2,3,4)
output = block(input)
outputoutput:
tensor([[[0.3116, 0.8282], [0.3116, 0.8282], [0.3116, 0.8282]], [[0.3116, 0.8282], [0.3116, 0.8282], [0.3116, 0.8282]]], grad_fn=<SigmoidBackward>)
Custom Modules Instead of using the predefined modules, we can also build our own by extending the nn.Module class. For example, we can build a the nn.Linear (which also extends nn.Module) on our own using the tensor introduced earlier! We can also build new, more complex modules, such as a custom neural network. You will be practicing these in the later assignment. To create a custom module, the first thing we have to do is to extend the nn.Module. We can then initialize our parameters in the __init__ function, starting with a call to the __init__ function of the super class. All the class attributes we define which are nn module objects are treated as parameters, which can be learned during the training. Tensors are not parameters, but they can be turned into parameters if they are wrapped in nn.Parameter class. All classes extending nn.Module are also expected to implement a forward(x) function, where x is a tensor. This is the function that is called when a parameter is passed to our module, such as in model(x).
class MultilayerPerceptron(nn.Module):
def __init__(self, input_size, hidden_size):
# Call to the __init__ function of the super class
super(MultilayerPerceptron, self).__init__()
# Bookkeeping: Saving the initialization parameters
self.input_size = input_size
self.hidden_size = hidden_size
# Defining of our model
# There isn't anything specific about the naming of `self.model`. It could
# be something arbitrary.
self.model = nn.Sequential(
nn.Linear(self.input_size, self.hidden_size),
nn.ReLU(),
nn.Linear(self.hidden_size, self.input_size),
nn.Sigmoid()
)
def forward(self, x):
output = self.model(x)
return outputHere is an alternative way to define the same class. You can see that we can replace nn.Sequential by defining the individual layers in the __init__ method and connecting the in the forward method.
class MultilayerPerceptron(nn.Module):
def __init__(self, input_size, hidden_size):
# Call to the __init__ function of the super class
super(MultilayerPerceptron, self).__init__()
# Bookkeeping: Saving the initialization parameters
self.input_size = input_size
self.hidden_size = hidden_size
# Defining of our layers
self.linear = nn.Linear(self.input_size, self.hidden_size)
self.relu = nn.ReLU()
self.linear2 = nn.Linear(self.hidden_size, self.input_size)
self.sigmoid = nn.Sigmoid()
def forward(self, x):
linear = self.linear(x)
relu = self.relu(linear)
linear2 = self.linear2(relu)
output = self.sigmoid(linear2)
return outputNow that we have defined our class, we can instantiate it and see what it does.
# Make a sample input
input = torch.randn(2, 5)
# Create our model
model = MultilayerPerceptron(5, 3)
# Pass our input through our model
model(input)output:
tensor([[0.6960, 0.5888, 0.6302, 0.5337, 0.6120],
[0.6787, 0.5964, 0.6672, 0.4974, 0.6041]], grad_fn=<SigmoidBackward>)We can inspect the parameters of our model with named_parameters() and parameters() methods.
list(model.named_parameters())output:
[('linear.weight', Parameter containing:
tensor([[-0.0094, -0.3072, 0.2230, 0.0499, -0.0917],
[ 0.0116, -0.2261, -0.4170, -0.1688, 0.2925],
[ 0.4049, 0.2189, 0.1391, 0.2115, -0.3926]], requires_grad=True)),
('linear.bias', Parameter containing:
tensor([0.1696, 0.2785, 0.3635], requires_grad=True)),
('linear2.weight', Parameter containing:
tensor([[ 0.4921, 0.5605, 0.5188],
[ 0.4088, 0.4430, 0.0042],
[-0.2919, 0.2893, -0.4794],
[ 0.4321, -0.1348, 0.4558],
[-0.4387, 0.2400, 0.3511]], requires_grad=True)),
('linear2.bias', Parameter containing:
tensor([ 0.2369, -0.0131, 0.4319, 0.1126, 0.2039], requires_grad=True))]

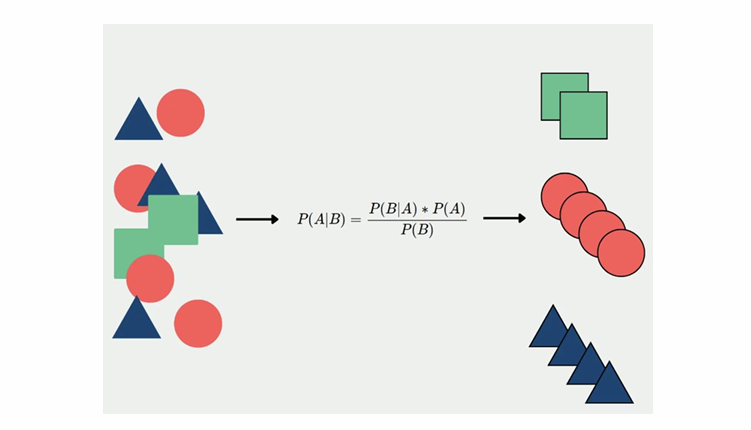
Comments